Our Research
Virtually every cell in our body has the same genetic information, yet individual cells can achieve highly specialized states with vastly differing functionality.
During development this is achieved through a series of cell expansion and differentiation cascades that allow initially pluripotent cells to commit to unique cells fates and enable complex multicellular development. Given that these outcomes are achieved with a fixed genetic complement (i.e. the same DNA in every cell), it reasons that specific cell types must invoke, with high fidelity, mechanisms that specify and restrict their capacity to express the correct complement of genes.
Learn more by watching Rob’s Francis Crick Prize Lecture at the Royal Society.
Controlling gene expression
It is clear that transcription factors which recognise specific DNA sequences in gene regulatory elements play a central role in defining gene expression outcomes. However, the vertebrate genome is vast, containing all the coding and structural information required to make gene products and effectively segregate chromosomes following cell division. Therefore, only a small percentage of the total genome sequence is utilized to directly specify whether or not genes should be expressed. Recognising this small fraction of the genome and then precisely controlling gene activity represents a formidable task for the cell’s gene regulatory machinery.
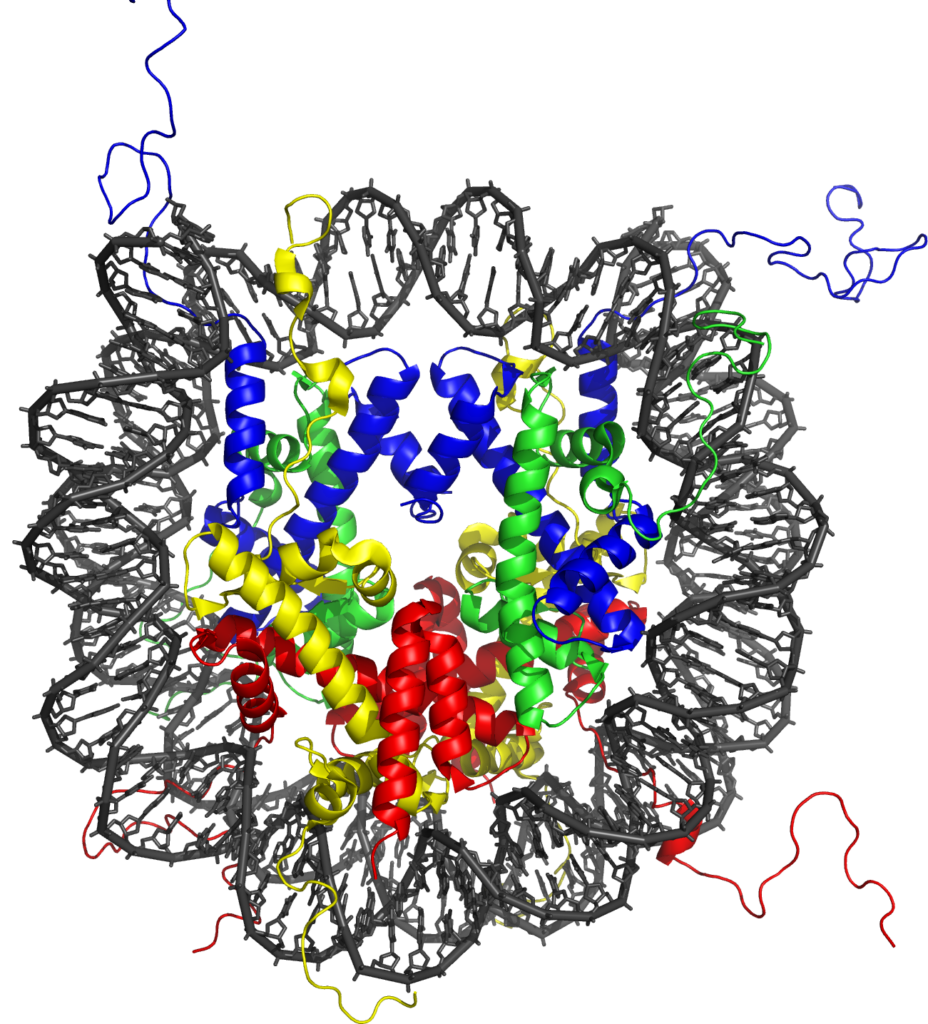
This task is complicated by the fact that DNA is not a naked template inside the cell, but is instead packaged into a structural entity called chromatin. Chromatin consists of a repeated subunit called the nucleosome, which contains DNA wrapped around protein molecules called histones. Recently it has become clear that gene regulatory elements in vertebrate genomes have very specific chemical modification signatures on their nucleosomes which differentiate them from surrounding non-regulatory regions. These chromatin states are thought to help highlight the location of gene regulatory regions in the genome, but, perhaps more importantly, emerging evidence suggests they also have central roles in controlling gene expression. Furthermore, chromatin modification states at individual genes can vary significantly between different cell types, suggesting they may also be key to how specific cell types initiate and maintain defined gene expression patterns.
However, our understanding of the molecular logic that defines how distinct chromatin modification states are initiated at gene regulatory elements and how they then control gene expression remains enigmatic and a major conceptual gap in our understanding of how our DNA encoded gene information is regulated. The Klose lab aims to address these fundamental questions and use our discoveries to understand how these basic processes are hijacked to cause human disease.
Chromatin and epigenetics
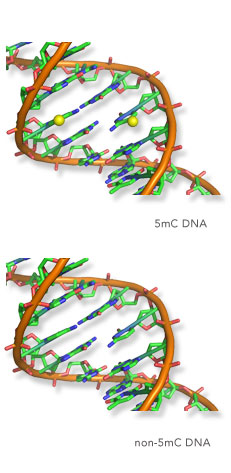
To uncover how chromatin modifications, or epigenetic modifications as they are often referred to, regulate gene expression, the Klose lab focusses on studying interesting regions of our genome which lack DNA methylation, called CpG islands. Importantly, CpG islands are intimately associated with gene promoters across the genome, but decades of research had failed to understand how or if these elements are required for controlling gene expression.
In addressing this important question, we discovered that CpG islands form a recognition site for a group of large molecular machines that can add or remove chemical modification to histones in chromatin (see our publications page for links to papers). We demonstrated that these CpG island-guided chromatin modifying systems can lead to the formation of either permissive (Trithorax) or repressive (Polycomb) chromatin states. These chromatin states are then fundamental to how cells are able maintain their normal gene expression programmes. If CpG island reading systems are disrupted, this causes developmental defects and other human diseases, including cancer.
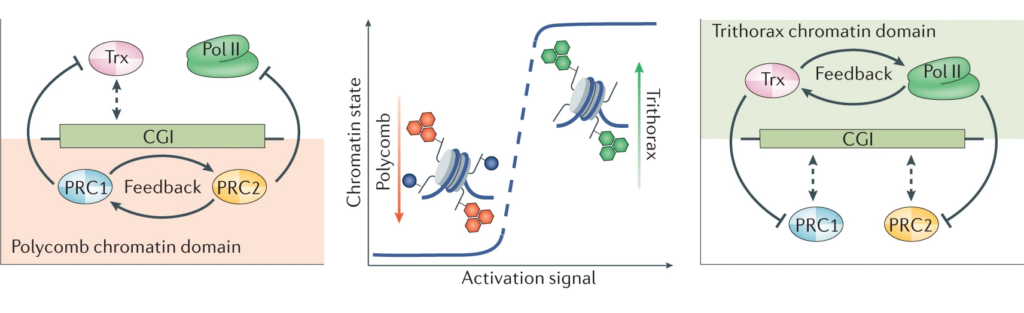
Building on our fundamental advances in understanding CpG island function, the lab is now focussed on tackling the next most pressing questions which include:
1. Discovering mechanistically how CpG island chromatin states control transcription to regulate gene expression.
2. Discovering how permissive and repressive chromatin states are integrated at CpG islands to control gene expression dynamics during cell lineage commitment and development.
3. Uncovering how perturbation of CpG island dependent gene regulatory systems leads to the molecular pathologies that cause disease.
We tackle these important problems by integrating a multidisciplinary yet complementary set of cutting-edge technologies spanning biochemistry, genome engineering, live-cell imaging, genomics, computational biology, and modelling. Through understanding how chromatin and epigenetic processes control gene expression, we hope in the long term to inform new therapeutic approaches to regulate these processes when they malfunction in cancer and other human diseases.
